BIOPHYS - Theoretical Physics tools applied to systems of biological interest
The BIOPHYS initiative focuses on the study of biological matter using mathematical modeling and computational methodologies of theoretical physics, using approaches which were originally developed in the context of the study of fundamental interactions.
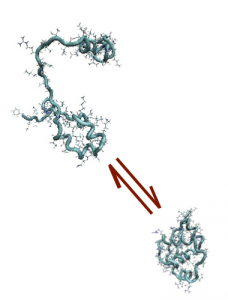
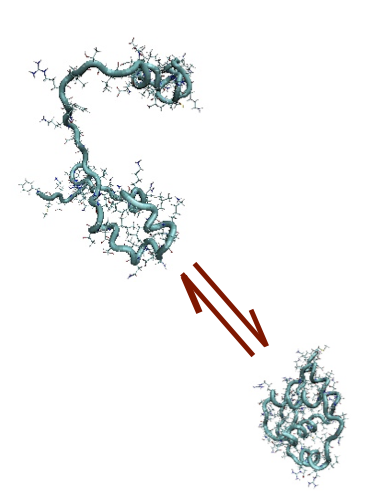
In particular, at TIFPA we study the physical mechanisms which underlay the protein folding, the conformational change through which proteins assume their well defined biologically active conformation (native state) starting from an initial coil-like configuration (see figure). This problem has relevant biomedical implications, since a number of severe pathologies emerge when the folding process goes wrong. We therefore explore the physical origin of misfolding processes.
We also study how electronic excitations, e.g. excited by light, propagate and dissipate in biomolecules. This study is important to understand photosynthesis and may have implications in the design of opto-electronic devices and solar cell.
The Science
The main goal of the BioPhys network is the study of problems and systems of Biological interest with the tools and ideas typical of theoretical physics. Our main interest is in Molecular Biology, a research context which has seen an impressive development in these last years and is nowadays of central relevance, both for the deep scientific questions which it raises and for the large number of potential biomedical applications. In this framework, our research project is organized in three main directions:
- Numerical simulations of systems of biological interest. (protein folding, biomolecules aggregation, protein-protein interactions...)
- Neural Networks and related systems. (Study of Brain connectivity, emergence of spatio-temporal patterns....)
- Regulatory Networks (Gene Regulatory Networks, Immune Networks, Chromatin organization....)
The common theme and the unifying setting of these research lines is the use of theoretical physics methods to model and analyse these problems. In particular methods borrowed from the statistical mechanics of disordered systems (Spin Glasses, Hopfield model...); computational tools typical of the simulation of many-body physical systems (Montecarlo Simulations, Molecular Dynamics,...); advanced mathematical tools (Graph theory, Network Theory, Stochastic Equations....). The most relevant aspect of our research network is the strong interaction of all the units with biologist. All the problems that we address stem from precise biological questions to which we try to give quantitative and testable answers. The research network is composed by ten nodes and involves more than 40 researchers.
The research activity of the Trento group focuses on the study of the non-equilibrium dynamics of classical and quantum excitations inside biomolecular systems. In this context, we develop and apply theoretical/computational enhanced sampling methods to investigate in atomistic detail rare conformational reactions, such as e.g. protein folding and mis-folding. We also develop non-equilibrium quantum field theory methods to study the transport of charged and neutral quantum excitations of the molecule electronic structure, in organic polymers and biological compounds.
TEAM
• INFN groups: Bari, Milano , Napoli, Pisa, Parma, Roma Tor Vergata, Salerno, TIFPA, Torino
• Principal Investigator: Michele Caselle (Torino)
• INFN Project: CSN IV
• Duration: n/a
TIFPA Team
• Local responsible for TIFPA: Pietro Faccioli
• Involved TIFPA people: Gianluca Lattanzi, Raffaello Potestio, Luca Tubiana, Giovanni Garberoglio, Roberto Menichetti, Thomas Tarenzi, Raffaele Fiorentini, Danial Ghamari, Marco Giulini, Giovanni Mattiotti, Lorenzo Petrolli, Marta Rigoli, Luigi Zanovello
Images